Table of Contents
Overview – Antigen Receptors
Antigen receptors are membrane-bound proteins found on B and T lymphocytes that enable the adaptive immune system to recognise and respond to a vast array of pathogens. These receptors — B cell receptors (BCRs) and T cell receptors (TCRs) — are genetically programmed during early development to bind specific antigens. Their diversity, generated through gene rearrangement and mutation, ensures immune adaptability. Understanding antigen receptors is foundational to immunology, vaccine responses, and immune tolerance.
Definition
- Antigen receptors = membrane-bound proteins on lymphocytes that bind specific antigens
- Include:
- BCRs (B cell receptors) = surface immunoglobulin molecules
- TCRs (T cell receptors) = heterodimeric proteins specific for peptide-MHC complexes
- Expression occurs before antigen exposure, during lymphocyte maturation in primary lymphoid organs
- Specificity is determined genetically, not environmentally
Function
- Detect presence of specific antigens in the environment
- Trigger clonal expansion, differentiation, and effector functions
- Each receptor is specific to one antigen (monospecific)
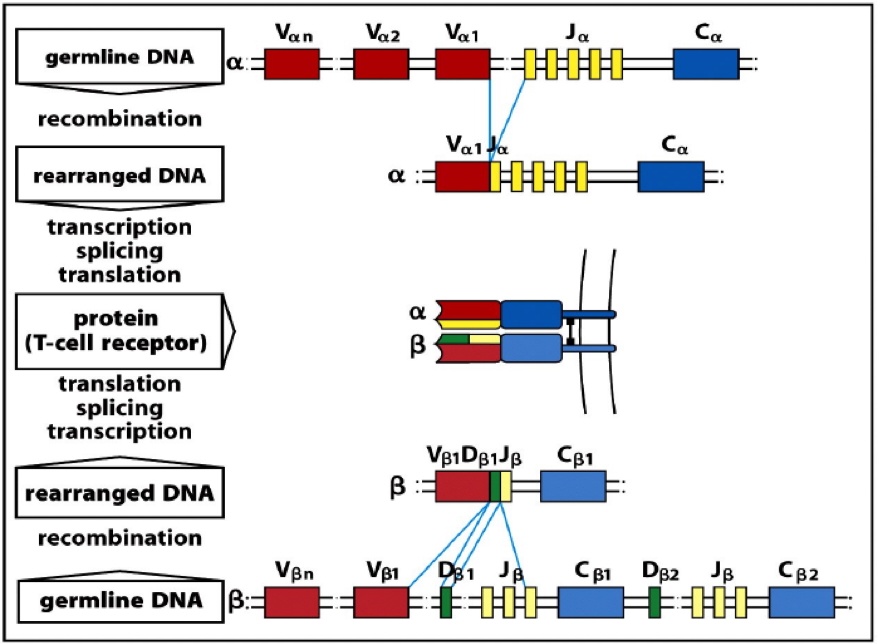
Generation of Diversity
Gene Rearrangement (in Primary Lymphoid Organs)
- Random recombination of gene segments during B and T cell development
- Heavy/β chains: V (variable), D (diversity), and J (joining)
- Light/α chains: V and J only
- Key enzymes:
- RAG-1 & RAG-2 recombinases
- Ligases
- Generates millions of unique antigen receptors

B cell-specific mechanisms (in Secondary Lymphoid Organs)
- Somatic Hypermutation (Affinity Maturation)
- Point mutations in variable region genes → higher/lower affinity
- High-affinity B cells are selected for survival
- Isotype Switching
- IgM constant region genes replaced with IgG/IgA/IgE
- CD4+ T helper cells required (via CD40/CD40L interaction)
- Alters antibody function but not specificity
B Cell Receptors (BCRs)
Morphology
- Membrane-bound antibodies (IgM or IgD in naïve cells)
- Structure:
- 2x heavy chains + 2x light chains
- Each chain has variable (V) and constant (C) regions
Functional Regions
- Fab (Variable Region):
- Binds intact antigens directly (proteins, carbohydrates, etc.)
- Contains complementarity-determining regions (CDRs)
- Fc (Constant Region):
- Signal transduction
- Antigen internalisation for MHC-II presentation
Binding Specifics
- Recognise whole antigens (unprocessed)
- Antigen binds via non-covalent forces:
- Electrostatic, hydrogen bonding, van der Waals, hydrophobic interactions
Diversity Generation
- In bone marrow: Ig gene rearrangement
- In lymph nodes/spleen:
- Somatic hypermutation
- Isotype switching


T Cell Receptors (TCRs)
Morphology
- Membrane-bound heterodimers
- Most T cells: αβ TCRs → CD4+ (helper) or CD8+ (cytotoxic)
- Minor subset: γδ TCRs → tissue-resident, innate-like function
- Structure:
- Each chain has separate V and C regions
Functional Regions
- Fab (Variable Region):
- Recognises processed peptides presented on MHC molecules
- Fc (Constant Region):
- Transduces intracellular signals upon antigen binding
Binding Specifics
- αβ T cells: Recognise antigen only when presented on MHC I/II
- γδ T cells: Can recognise whole antigen (no MHC restriction)
Diversity Generation
- Occurs in thymus via gene rearrangement
- No isotype switching or somatic hypermutation


Summary – Antigen Receptors
Antigen receptors are genetically encoded structures on B and T cells that detect specific antigens and initiate immune responses. Through gene rearrangement and, in B cells, further refinement via somatic hypermutation and isotype switching, the immune system achieves vast specificity. For a broader context, see our Immune & Rheumatology Overview page.